BIV: Batch Inverse Variance Weighting Deep Heteroscedastic Regression Using Privileged Information
Introduction
The performance of deep supervised learning methods is impacted when the training dataset, on which the parameters are optimized, and the testing dataset, which evaluates the performance of the model on the task, are not sampled from identical distributions. In heteroscedastic regression, the label for each training sample is corrupted by noise coming from a different distribution. In some cases, it is possible to know an estimate of the variance of the noise for each label, which quantifies how much it contributes to the misalignment between the datasets. We propose an approach to include this privileged information in the loss function together with dataset statistics inferred from the mini-batch to mitigate the impact of the dataset misalignment. We adapt the idea of Fisher-information weighted average to function approximation and propose Batch Inverse-Variance weighting. We show the validity of this approach as it achieves a significant improvement of the performances of the network when confronted to high, input-independent noise.
Prerequisites
To run the code, we wrapped up all the used libraries inside a singularity container, you can download it here. To manually build your environment using anaconda, we provide the yml file here.
Run the Code
To run the code locally:
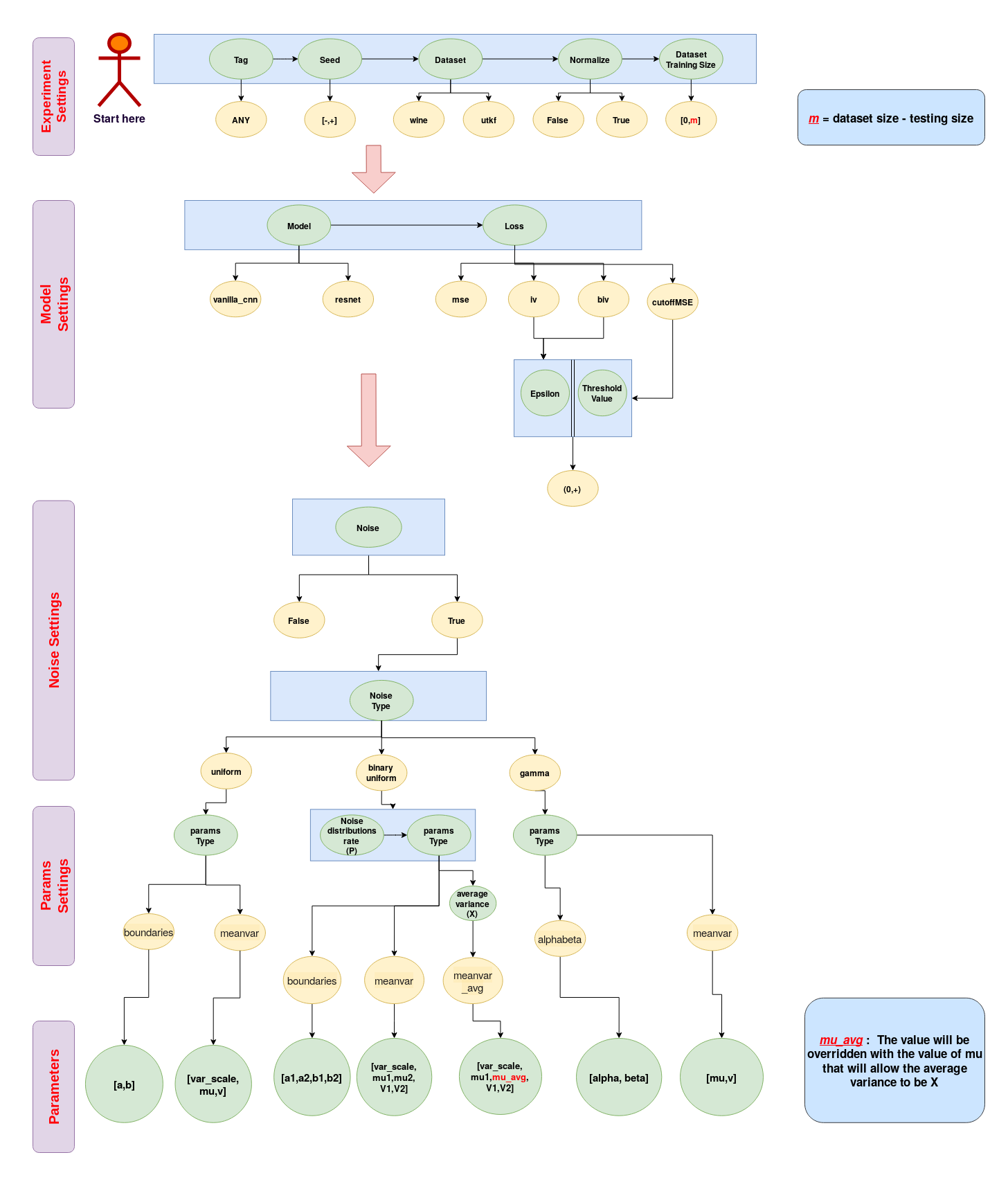
python main.py --experiment_settings="exp_tag,7159,utkf,True,16000" --model_settings="vanilla_cnn,cutoffMSE"
--noise_settings="True,binary_uniform" --params_settings="meanvar_avg,0.5,3000" --parameters="True,0.5,1,0.3,0"
To run the code locally inside singularity container:
singularity exec --nv -H $HOME:/home/ -B ./your_dataset_directory:/datasets/ -B ./your_outputs_directory:/final_outps/ ./your_environments_directory/pytorch_f.simg python /path/to/main.py --experiment_settings="exp_tag,7159,utkf,True,16000"
--model_settings="vanilla_cnn,cutoffMSE" --noise_settings="True,binary_uniform" --params_settings="meanvar_avg,0.5,3000"
--parameters="True,0.5,1,0.3,0"
To run the code in a cluster that supporting slurm workload manager, use this starter script:
#!/bin/bash
#SBATCH -o /path/to/logs/noise_%j.out # Change this!
#SBATCH --cpus-per-task=4
#SBATCH --gres=gpu:1
#SBATCH --mem=32Gb
# Load cuda (it is not needed if have it enabled as a default.)
module load cuda/10.0
# 1. You have to load singularity (it is not needed if have it enabled as a default.)
module load singularity
# 2. Then you copy the container to the local disk
rsync -avz /path/to/pytorch_f.simg $SLURM_TMPDIR # Change this!
# 3. Copy your dataset on the compute node
rsync -avz /path/to/your_dataset/ $SLURM_TMPDIR # Change this!
# 3.1 export wandb api key
export WANDB_API_KEY="put your wandb key here" # Change this!
# 4. Executing your code with singularity
singularity exec --nv -H $HOME:/home/ -B $SLURM_TMPDIR:/datasets/ -B $SLURM_TMPDIR:/final_outps/ $SLURM_TMPDIR/pytorch_f.simg python /path/to/main.py --experiment_settings=$1 --model_settings=$2 --noise_settings=$3 --params_settings=${4-"None"} --parameters=${5-"None"}
# 5. Move results back to the login node.
rsync -avz $SLURM_TMPDIR --exclude="your_dataset" --exclude="pytorch_f.simg" /path/to/outputs # Change this!
# Note:
# $SLURM_TMPDIR = The compute node directory.
then run the script with sbatch:
sbatch --gres=gpu:rtx8000:1 ./path/to/main.sh "exp_tag,7159,utkf,True,16000" "vanilla_cnn,cutoffMSE" "True,binary_uniform" "meanvar_avg,0.5,3000" "True,0.5,1,0.3,0"
Examples
-
To run a vanilla CNN while normalising the data, where the loss function is MSE:
python main.py --experiment_settings="exp_tag,7159,utkf,True,16000" --model_settings="vanilla_cnn,mse,0.5" --noise_settings="False" -
To run resnet-18 with BIV loss (epsilon=0.5), where the noise variance is coming from a single uniform distribution:
python main.py --experiment_settings="exp_tag,7159,utkf,True,16000" --model_settings="resnet,biv,0.5" --noise_settings="True,uniform" --params_settings="boundaries" --parameters="0,1" -
To run resnet-18 with BIV loss (epsilon=0.5), where the noise variance is coming from a single uniform distribution that has a variance equal to the maximum heteroscedasticity:
python main.py --experiment_settings="exp_tag,7159,utkf,True,16000" --model_settings="resnet,biv,0.5" --noise_settings="True,uniform" --params_settings="meanvar" --parameters="True,0.5,0.083" -
To run resnet-18 with BIV loss (epsilon=0.5), where the noise variance is coming from a bi-model (uniform) distribution, in which the weight of the contribution of the both distributions is equal (0.5):
python main.py --experiment_settings="exp_tag,7159,utkf,True,16000" --model_settings="resnet,biv,0.5" --noise_settings="True,binary_uniform" --params_settings="boundaries,0.5" --parameters="0,1,1,4" -
To run resnet-18 with MSE loss, where the noise variance is coming from a bi-model (uniform) distribution by specifying the mean and variance of this model:
python main.py --experiment_settings="exp_tag,7159,utkf,True,16000" --model_settings="resnet,mse" --noise_settings="True,binary_uniform" --params_settings="meanvar,0.5" --parameters="False,0.5,1,0.083,0" -
To run resnet-18 with BIV loss (epsilon=0.5), where the noise variance is coming from a bi-model (uniform) distribution in which the average mean is 2000.
python main.py --experiment_settings="exp_tag,7159,utkf,True,16000" --model_settings="resnet,biv,0.5" --noise_settings="True,binary_uniform" --params_settings="meanvar_avg,0.5,2000" --parameters="False,0.5,1,0.083,0" -
To run resnet-18 with MSE loss, where the noise variance is coming from a bi-model (uniform) distribution and with noise threshold=1:
python main.py --experiment_settings="exp_tag,7159,utkf,True,16000" --model_settings="resnet,cutoffMSE,1" --noise_settings="True,binary_uniform" --params_settings="meanvar_avg,0.5,2000" --parameters="False,0.5,1,0.08,0"
Command-line Arguments: Flow Chart